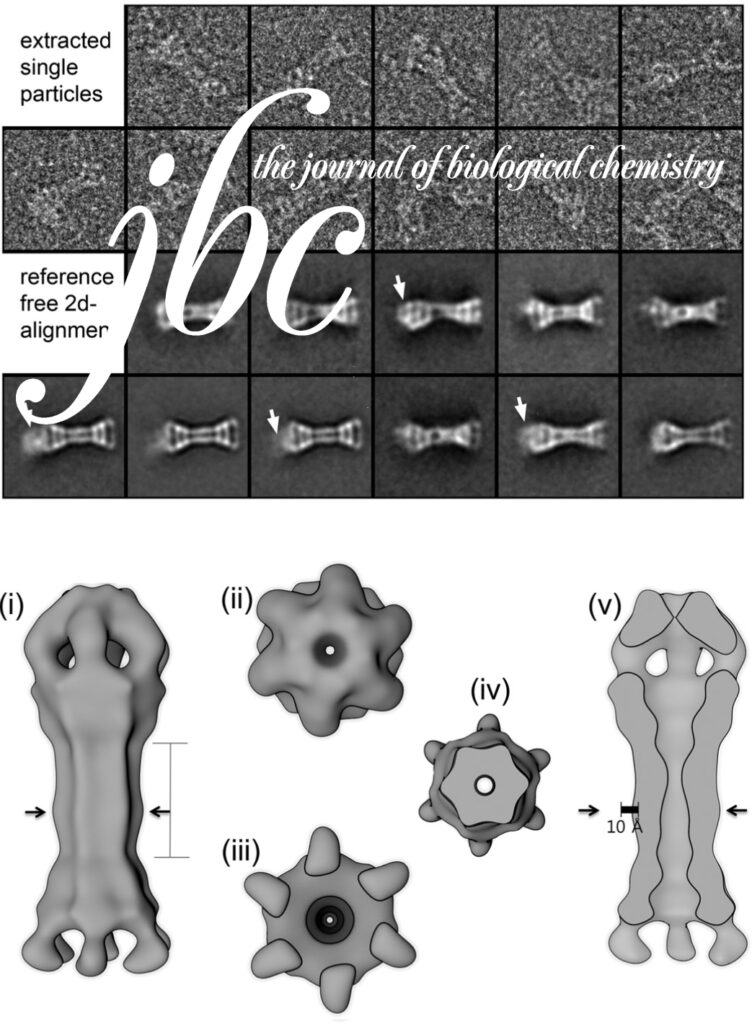
Nucleotide-dependent conformational changes in the N-Ethylmaleimide Sensitive Factor (NSF) and their potential role in SNARE complex disassembly.
Homohexameric, N-Ethylmaleimide Sensitive Factor (NSF) disassembles Soluble NSF Attachment Protein Receptor (SNARE) complexes after membrane fusion, an essential step in vesicular trafficking. NSF contains three domains (NSF-N, NSF-D1, and NSF-D2), each contributing to activity. We combined electron microscopic (EM) analysis, analytical ultracentrifugation (AU) and functional mutagenesis to visualize NSF’s ATPase cycle.