Roman Levin, Zoe Köck, Janosch Martin, René Zangl, Theresa Gewering, Leah Schüler, Arne Moeller, Volker Dötsch, Nina Morgner, Frank Bernhard
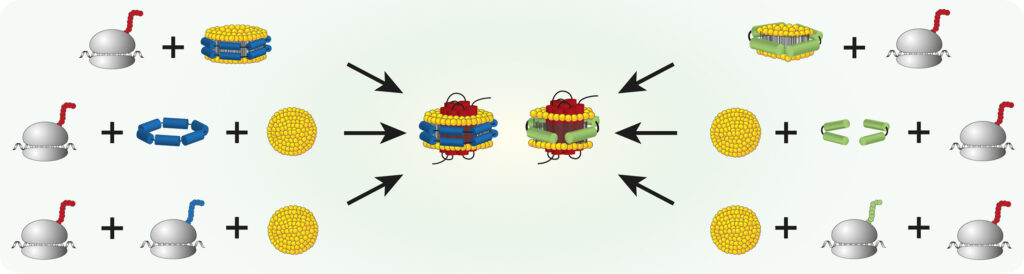
Nanoparticles composed of amphiphilic scaffold proteins and small lipid bilayers are valuable tools for reconstitution and subsequent functional and structural characterization of membrane proteins. In combination with cell-free protein production systems, nanoparticles can be used to cotranslationally and translocon independently insert membrane proteins into tailored lipid environments. This strategy enables rapid generation of protein/nanoparticle complexes by avoiding detergent contact of nascent membrane proteins. Frequently in use are nanoparticles assembled with engineered derivatives of either the membrane scaffold protein (MSP) or the Saposin A (SapA) scaffold. Furthermore, several strategies for the formation of membrane protein/nanoparticle complexes in cell-free reactions exist. However, it is unknown how these strategies affect functional folding, oligomeric assembly and membrane insertion efficiency of cell-free synthesized membrane proteins. We systematically studied membrane protein insertion efficiency and sample quality of cell-free synthesized proteorhodopsin (PR) which was cotranslationally inserted in MSP and SapA based nanoparticles. Three possible PR/nanoparticle formation strategies were analyzed: (i) PR integration into supplied preassembled nanoparticles, (ii) coassembly of nanoparticles from supplied scaffold proteins and lipids upon PR expression, and (iii) coexpression of scaffold proteins together with PR in presence of supplied lipids. Yield, homogeneity as well as the formation of higher PR oligomeric complexes from samples generated by the three strategies were analyzed. Conditions found optimal for PR were applied for the synthesis of a G-protein coupled receptor. The study gives a comprehensive guideline for the rapid synthesis of membrane protein/nanoparticle samples by different processes and identifies key parameters to modulate sample yield and quality.
DOI: 10.1016/j.bbamem.2022.184017
PMID: 35921875